Failing Hard: How I 'lost' two years of data
Failure is one of those topics that is discussed far less than it should be ---It is hard to tell other people about mistakes you have made--- yet failure is usually a far better teacher than success. This is why I want to share my story of how I spent the first two years of my PhD collecting useless data because of a bug in my code.
The Problem
I perform computer simulations of toy molecular systems, using something simple and understandable to gain insights into more complicated chemical systems. This is just like in High School and Undergraduate Physics where you perform calculations for objects in a frictionless vacuum. Apart from the simplicity of a toy system, a major benefit of using one is that you can modify individual parameters of the system to test a hypothesis. I had the hypothesis that rotational motion plays an important role in the large scale translational motion---known as diffusion, so I was using a toy system which allowed me to control the effective magnitudes of these two types of motion. The small scale translational motion is related to the mass; heavy objects are more difficult to move in a straight line. While the rotational motion is related to the moment-of-inertia, a measure of how difficult something is to rotate. Though varying these two properties, the mass and the moment-of-inertia, I can establish how the translational and rotational motions contribute to the rate of diffusion. Setting the moment-of-inertia really large effectively stops the rotational motion, so what happens to the diffusion? Or the opposite, making the moment-of-inertia really small what is the resulting effect on the diffusion?
Before I start changing the parameters of the toy model, I need a reference (or control) model related to existing results. Science is always performed with reference to our current understanding, so the control model provides a point of comparison to existing results, including other toy models and real chemical systems. The molecule I am studying is comprised of three circles, one large and two small, arranged like a Mickey Mouse head.
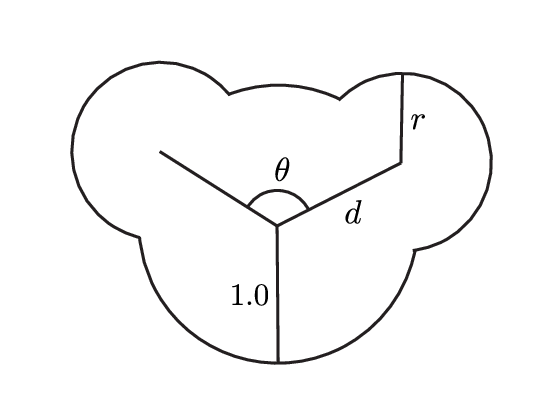
To make calculations with these toy systems easier, I use a system of units in which most quantities have a unitless value of 1. Each particle has a point mass at the center of the circle of 1, the large circle has a radius of 1, a reasonable temperature is 1 ---It is really nice that most of the multiplication and division disappears. The smaller particles in the molecule have a radius of $0.63$ and are located on the circumference of the large particle at an angle of $120^\circ$.
I am using the above molecule in Molecular Dynamics (MD) simulations, which use Newton's equations of motion to update the positions of each particle. The translational acceleration $a$ of a molecule is related to the force $F$ acting on it divided by the mass $m$ as given by the equation $a=F/m$. While the angular acceleration $\alpha$ is related to the torque $\tau$ divided by the angular momentum $I$ as given by $\alpha = \tau/I$. Molecular Dynamics simulations have two different methods of dealing with the molecules I am using. The first is to solve these equations for each of a molecule's component particles individually, followed by a second step which restores the molecular shape. The second approach is to calculate the force on each component particle, then adding them together to solve the above equations for the molecule as a whole.
Molecular Dynamics simulations are a standard computational tool for understanding a variety of problems from crystal growth to protein folding. As a widely used tool, there are many different software packages available for performing these simulations, with each having their own strengths and weaknesses. Using these software packages involves expressing the simulation you want to perform in the appropriate manner for the software. At the start of my PhD I decided that Hoomd developed by the Glotzer group at the University of Michigan was the most suitable for the types of simulations I was going to be performing. Soon after starting to understand the software, there was a new major release, which included a significant overhaul of the software. This overhaul included changing how it handled the molecular calculations from the first method to the second method, something which I didn't realise at the time. In getting my simulations to work with the new version of Hoomd, I manually entered a moment of inertia, calculated from each particle having a point mass of 1, however the total mass of the molecule was taken as the mass of an individual particle being 1 instead of 3. By not actually checking the simulations were behaving as I intended I spent the next two years of my PhD characterising the behaviour of the wrong control experiment.
Finding the Bug
I found the bug as part of trying to add additional functionality. I was trying to randomly initialise velocities of particles in a configuration such that they matched the desired temperature.
The reason I found the bug was a result few the Bug result of setting thermodynamic quantities - temperature is a distribution of velocities - energy is evenly distributed between translational and rotational motion
Lessons Learnt
The lessons I have learnt from this experience can be grouped into two groups, the identification of bugs, and the management of computational projects.
Identifying Bugs
Even before the identification of this bug I had developed a test suite for my code, ensuring I was configuring the simulations in Hoomd with the appropriate values. What I wasn't checking, was that these values produced the simulations I thought I was running. Typically the method of doing this is to take some previous results and replicating them, although since no-one else has run simulations on the particular system I am using it is a little difficult to do this.
I had been told by my supervisor on many occasions to confirm that the simulations were physically accurate, usually with reference to conservation of energy. This was really easy to ignore because that out of scope for the code I was writing, however, the idea of checking the simulation is consistent with the laws of physics was definitely worth considering earlier.
Testing is not about ensuring exact correctness, it is also ensuring the absence of obvious errors.
A simple check of physicality is though the equi partition theorem, which describes that each of the degrees of freedom, whether translational or rotational, have the same energy. The relationships below link the mass $m$ and moment-of-inertia $I$ to the temperature $T$ providing a route to testing the simulations, where the angled brackets $\langle \rangle$ denote an averaged quantity.
What might not be so obvious here is that the temperature $T$ in the above equations is not a static quantity, instead fluctuating about the desired temperature, a construct of the type of simulations I am performing. This makes the task of testing the temperature is correct significantly more difficult, since it is not a set value, lying in a distribution. While creating automated testing for whether a value that varies can be difficult, re-framing the test as not-wrong is just as valid. With this bug I created I was off by a factor of 3, many times larger than any reasonable variation. I didn't need to test that the temperature was exactly right, I just needed to ensure it is not obviously wrong.
Rebuilding the Dataset
While the main storyline of this article has been losing the years of my PhD research, a major part of the learning experience has been the rebuilding of all the data I had collected.
Reproducibility in science is not just for others to reproduce your work it is also so you can reproduce it when you stuff up.
I have been constantly working to improve my workflow, as I learn more about what works and what doesn't ending up with a structure based heavily on the cookiecutter-cms and cookiecutter-data-science project templates. All the new work was structured into these project templates which are well organised, with an emphasis on being able to replicate analyses. What I really noticed in rebuilding my dataset was that replicating these newer elements was simple and straightforward, while the older projects just working out what I had done was a challenge, let alone what needed to be recreated.
Further to having a well organised data analysis, having all the computational experiments, including the permutations of every variable and the scripts required to run the simulation was invaluable. The experiments where this was up to date were the simplest to deal with, I just needed to run them again.
Conclusion
Although we often don't like to admit it, we all make mistakes. Mistakes, failures, and bugs are all part of doing research. While they may be disruptive and inconvenient, they are only truly damaging when they keep re-occurring. Making a mistake once is part of the process, repeatedly making the same mistake is a problem. Best practices like automated testing are ways of making the mistakes that do occur obvious and simple to fix, as well as providing a way of preventing them from reoccurring. I am now fairly confident that should this particular bug reappear I will notice, although that doesn't mean there isn't another hiding away for me to find.